Train a model with weight sparsity#
Overview#
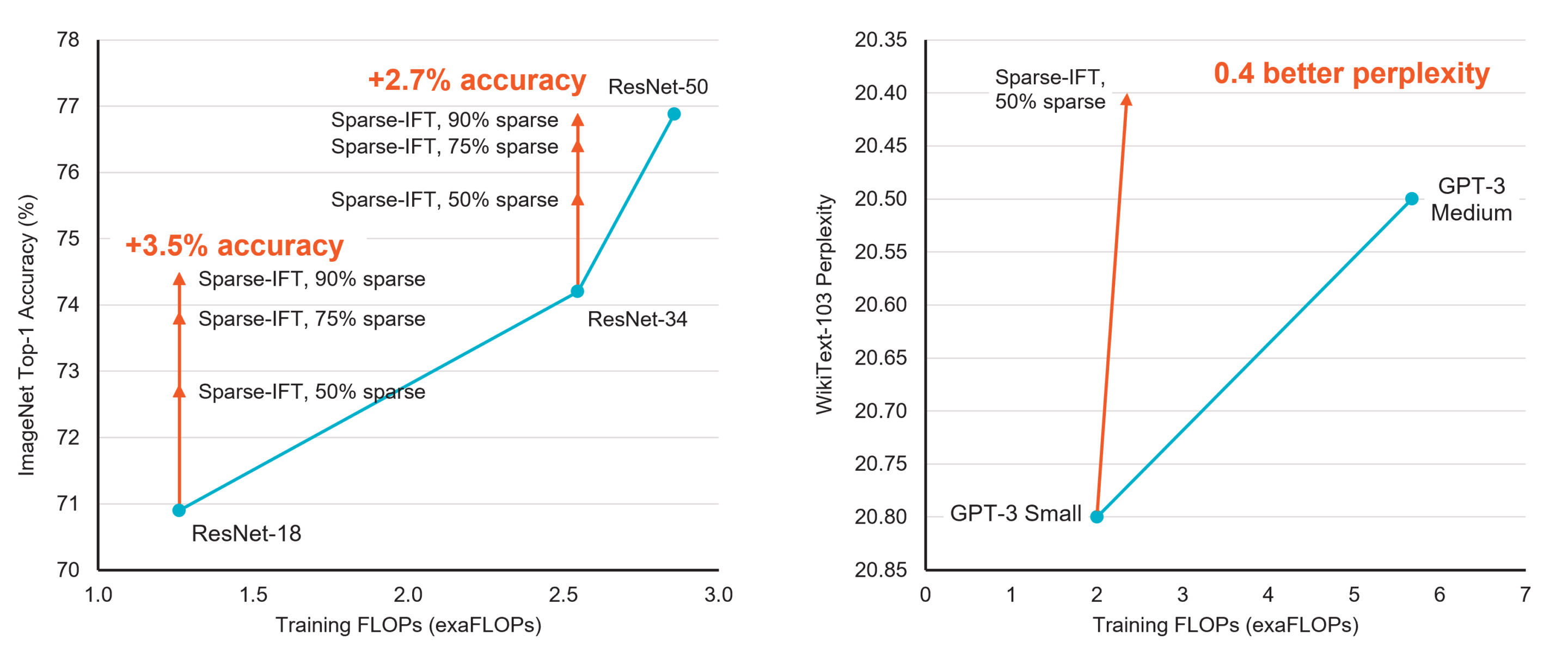
In 2018, state-of-the-art neural networks such as BERT had a few hundred million parameters. Two years later, the world was introduced to GPT-3. With 175 billion parameters and a 3.14*1023 FLOPs (floating point operations) compute budget, it is estimated to have required 10,000 NVIDIA V100 GPUs for 15 days, accounting for 552 tons of CO2e emissions and 1,287 MWh of energy [Patterson et al.].
While model sizes continue to increase in the pursuit of better accuracy, the resulting compute and memory requirements make these models intractable for most practitioners. When coupled with hardware that accelerates unstructured sparsity, weight sparsity is a promising way to train models using significantly less computing and memory.
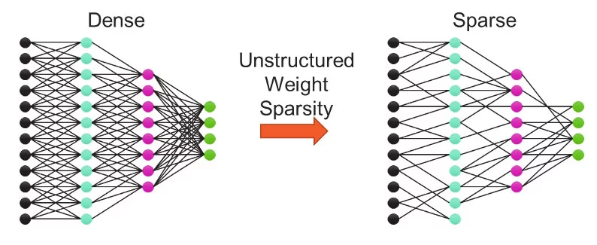
Weight sparse training methods set subsets of weights to zero during training, often the ones already close to zero in magnitude. The resulting sparse model requires far fewer FLOPs to train and fewer parameters to store, as multiplies with zeros get skipped on both forward and backward passes through the network. Only systems that can accelerate sparsity, such as Cerebras CS-2, can take advantage of the lower resource requirement and use the reduction in FLOPs to accelerate training significantly. Finding and training sparse models to match the accuracy of their original “dense” (i.e., non-sparse) configurations is an active and open area of research! For software release 1.8, we are exposing an early preview static sparsity mechanism to allow experimentation with this exciting capability.
Sparsity via YAML#
When using the Cerebras Model Zoo reference models, runs are parameterized via
YAML configs that include model, data, and optimizer parameters (GPT-3 example
here). To
train with sparsity, you can include a sparsity section in your run’s YAML
config file as a sibling to the model and optimizer sections. Each
parameter is sparsified independently. i.e. we don’t yet support global
sparsity.
For example, with the following config, the sparsity level is set to 0.3 (30%),
and init_method is "random", which means 30% of weights for each layer
will be pruned once at model initialization and kept that way throughout
training.
sparsity:
type: "static"
sparsity: 0.3
init_method: "random"
Sparsity is parameterized primarily by the following keys:
type: Algorithm or type of sparsity to apply.static(StaticSparsityOptimizer)gmp(GMPSparsityOptimizer)set(SETSparsityOptimizer)rigl(RigLSparsityOptimizer)name of a class inheriting from
DynamicSparsityOptimizerto allow you to use the same config-driven system to instantiate your own custom implementations.
init_method: Method to compute the initial sparsity distribution.random: (default) Sparsity is randomly distributed within each weight.topk: Sparsity is distributed according to the lowest magnitude weights.from_zeros: Weights with a zero value are treated as pruned, all others are treated as connected. 1
sparsity: The desired sparsity level between 0 and 1.0.0 means dense, 1.0 means all connections are pruned. Dynamic sparsity types also accept more complex configuration described below in Dynamic HyperParameters.
param_name_patternsoptional: Control which parameters are havesparsity applied. When this is omitted, any multidimensional weights (except those with
embedding,norm, orlm_headin their name) automatically get sparsity applied. That provides a good default heuristic for transformer based models 2, but otherwise a regular expression can be provided (e.g.param_name_patterns: "(.*dense_layer.weight)|(.*linear_layer.weight)") to only apply sparsity to parameters which match.This can also configure sparsity options per-layer when given as a dictionary described below in advanced param_name_patterns.
Dynamic Sparsity Schedule#
Dynamic sparsity (e.g. type: gmp, set, or rigl) needs an
additional schedule indicating when to update the sparsity pattern. There
are 2 basic methods built-in with 3 different options:
Regular Interval#
When sparsity should be updated at a regular interval, a single period can be given:
sparsity:
schedule: 100
type: set
sparsity: 0.9
To control beginning and ending steps, use a dictionary. In the following example, training will start dense, sparsity will be applied for the first time on step 77, and every 100 steps thereafter (177, 277, 377) and perform no more updates after step 377 (stop is exclusive):
sparsity:
schedule:
start: 77
freq: 100
stop: 477 # An update will _not_ be performed on step 477
type: set
sparsity: 0.9
Irregular Interval#
When sparsity should be updated at arbitrary steps, specify them in a list:
sparsity:
schedule: [0, 5, 20, 50]
type: set
sparsity: 0.9
Dynamic HyperParameters#
Dynamic sparsity (e.g. type: gmp, set, or rigl) can configure
the sparsity (and drop_fraction for set and rigl) field using a
“step aware hyperparemeter” akin to learning rate schedules in addition to
simple constants. These more complex configurations usually require additional
options and so are specified as dictionaries.
Note
The base
DynamicSparsityOptimizer that
invokes such a dynamic hyperparameter for sparsity ensures sparsity
levels stay legal by using torch.clamp(sparsity, min=0.0, max=1.0).
Linear#
\(y(step) = init + slope \cdot step\)
sparsity:
type: "gmp"
schedule: 1000
sparsity:
type: "linear"
init: 0.0 # value at step zero
slope: 0.001 # increase in value each step
Exponential#
\(y(step) = final + (init-final) e^{step \cdot gamma}\)
This is expecially useful for gmp, where the sparsity level monotonically
increases throughout training because a fraction of the remaining connections
are pruned at each update step, asymptotically approaching an empty network.
sparsity:
type: "gmp"
schedule: 1000
sparsity:
type: "exp"
init: 0.0 # starting value
final: 1.0 # asymptotic ending value
# Prune 10% of the remaining connections every 1000 steps
gamma: -0.00010536051 # ln(1-0.10)/1000
Cosine#
\(y(step) = o + a \cdot \cos(step \cdot \pi / half\_period)\), where \(o = (init + minimum)/2\) and \(a = init - o\).
This is especially useful for rigl, which usually uses a “cosine decay” on
its drop_fraction. minimum defaults to 0.0. half_period
controls what step the value reaches its minimum.
sparsity:
type: "rigl"
schedule: 1000
sparsity: 0.9
drop_fraction:
type: "cosine"
init: 0.3 # starting value
half_period: 10000 # reaches minimum (default 0) after 10 updates
More Config examples#
The most basic configuration, applying random 30% sparsity to all Dense layer weights:
sparsity:
type: "static"
sparsity: 0.3
Apply uniform (static) sparsity to a selected set of weights, with a sparsity pattern guided by the weight magnitudes:
sparsity:
type: "static"
sparsity: 0.9
init_method: "topk"
param_name_patterns:
- "dense_layer.weight"
- "linear_layer.weight"
Basic dynamic sparsity using the SET algorithm. Update the sparsity pattern every 1000 iterations.
sparsity:
type: "set"
sparsity: 0.9
schedule: 1000
drop_fraction: 0.3
Advanced param_name_patterns#
When each parameter (or group of parameters) needs different configuration,
param_name_patterns can be specified as a dictionary, mapping “patterns” to
the config dictionaries to overlay on the default sparsity config options.
For example, when using RigL on transformer networks (uses gradient information to guide which conections to prune), sparsity can be cyclically restributed between the heads of attention projection weights in case samples in a batch activate one head disproprtionetly to another. This ultimately decreases the effectiveness of dynamic sparsity and even can hurt model performance.
To ensure sparsity is fairly distributed between the different attention heads
of the the multi-head attention projections, you can specify
balance_out_groups when the _output_ logits are logically N
independent/stacked groups (i.e. input projection weights before multi-head
attention QKV), or balance_in_groups for the reverse (i.e. output
projection weights). These should apply differently to different weights using
param_name_patterns since this conceptually only applies to Attention
projection weights. In the following example, the model has 12 attention heads.
sparsity:
type: "rigl"
sparsity: 0.9
schedule: 1000
drop_fraction:
type: "cosine"
init: 0.3
half_period: 10000
param_name_patterns:
".*proj_[qkv]_dense_layer.weight":
balance_out_groups : 12 # ensure this matches model.num_heads
".*proj_output_dense_layer.weight":
balance_in_groups : 12 # ensure this matches model.num_heads
".*linear_layer.weight":
Note
The “value” in the above example for the last (.*linear_layer.weight*.)
key is nil (YAML) or None (Python). This means to just use the default
options specificied at the top level, but still select certain parameters via
regex)
Running a Sparse Model#
No change is needed to the run command (see guide: Launch your job) -
ensure the .yaml file has sparsity enabled. To validate your sparsity
config before launching training, run with --validate_only. You can also
log which weights are being sparsified by passing --logging VERBOSE to your
run command.
python modelzoo/path/model/run.py CSX \
--params params_with_sparsity.yaml \
--num_csx=1 \
--model_dir model_dir --mode {train,eval,eval_all,train_and_eval} \
--mount_dirs {paths modelzoo and to data} \
--python_paths {paths to modelzoo and other python code if used}
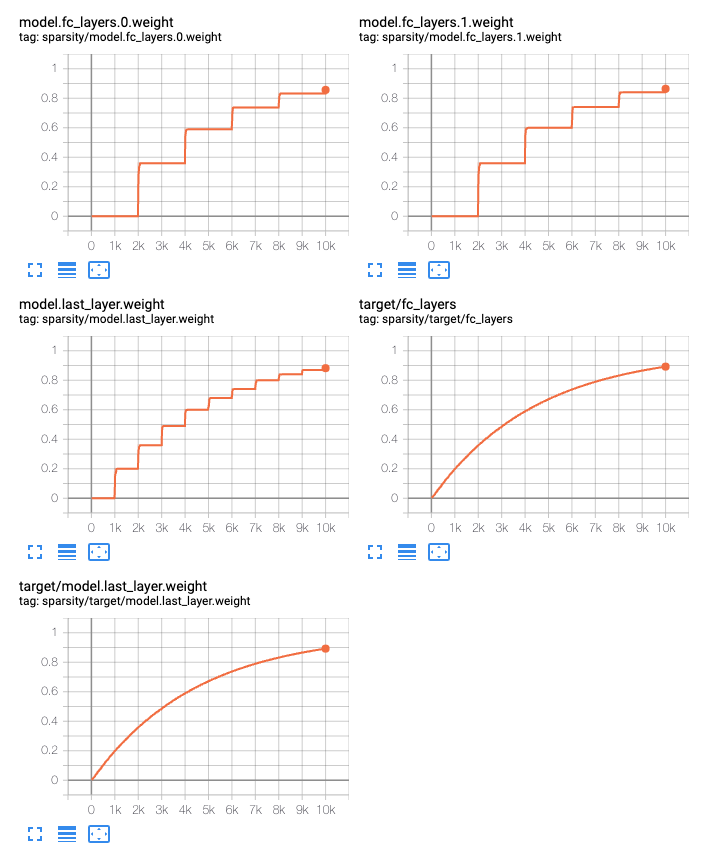
When using dynamic sparsity, you can see realtime summaries by setting the
config field add_summaries: True. Each parameter group independent
summarizes its target sparsity as well as the effective sparsity of each
tensor. To aide in debugging, groups can be given a name, otherwise one is
generated either from the name of the parameter if the group contains only a
single or a generic group_$N otherwise.
sparsity:
# Enable tensorboard summaries for target and actual sparsity
add_summaries: True
type: "gmp"
sparsity:
type: "exp"
init: 0.0 # starting value
final: 1.0 # asymptote value
# Prune 20% of the remaining connections every 1000 steps:
gamma: -0.0002231435513142097 # ln(1-0.20)/1000
param_name_patterns:
".*fc_layers.*weight":
# Name this group, just for debugging.
name: "fc_layers"
schedule: 2000
".*last_layer.*weight":
schedule: 1000
Sparsity via API#
If you provide your own training loop without using the run.py
infrastructure from modelzoo, you can construct and use a SparsityOptimizer
with a convenient API:
# Construct model and optimizer as usual
model = torch.nn.Module(...)
optimizer = cstorch.optim.SGD(...)
# Construct a sparsity optimizer, and use the returned wrapper as a drop-in
# replacement for the original optimizer
optimizer = cstorch.sparse.configure_sparsity_wrapper(
model,
optimizer,
sparsity_type="static",
sparsity=0.7,
init_method="topk",
)
# Model forward and backward as usual. Sparsity is automatically applied.
loss = model(...)
loss.backward()
# Update weights. If using dynamic sparsity, it is also updated according
# to its schedule.
optimizer.step()
See cerebras_pytorch.sparse for a description of the API methods such
as configure_sparsity_wrapper used for
configuring and using instances of the
BaseSparsityOptimizer. The helper
configuration methods all take **kwargs containing the Python versions of
the above YAMl configs, including sparsity, init_method, schedule,
param_name_patterns etc. However, when envoking the API directly, python
lambdas or user functions can be passed in for most hyper parameters, including:
init_method, sparsity (for dynamic sparsity), and schedule. The
signature of each function varies, so consult the Callable type hints in
the API reference.
SparsityWrapperOptimizer#
The above example is equivalent to:
# Construct model and optimizer as usual
model = torch.nn.Module(...)
optimizer = cstorch.optim.SGD(...)
# Construct a sparsity optimizer directly.
sparsity_optimizer = cstorch.sparse.configure_sparsity_optimizer(
sparsity_type="static",
model.named_parameters(),
sparsity=0.7,
init_method="topk",
)
# Install the "apply_sparsity" hooks so that the model's .forward()
# always computes using the sparse view of the parameters and the
# gradients are sparsifed before any use.
sparsity_optimizer.hook_module(model)
# Replace the optimizer with a wrapper so that sparsity state is saved,
# sparsity masks are updated by .step(), and conditional execution of
# dynamic gradient scaling handles skipping sparsity update too.
optimizer = cstorch.sparse.SparsityWrapperOptimizer(
optimizer, sparsity_optimizer
)
for step in dataloader:
# Model forward and backward as usual. Sparsity is automatically applied.
loss = model(...)
loss.backward()
# Update weights. If using dynamic sparsity, it is also updated according
# to its schedule.
optimizer.step()
SparsityOptimizer manual use#
The above example is also equivalent to:
# Construct model and optimizer as usual
model = torch.nn.Module(...)
optimizer = cstorch.optim.SGD(...)
# Construct a sparsity optimizer using configuration helper.
sparsity_optimizer = cstorch.sparse.configure_sparsity_optimizer(
sparsity_type="static",
model.named_parameters(),
sparsity=0.7,
init_method="topk",
)
for step in dataloader:
# Ensure weights state are sparse. Also sets up gradient
# hook to apply sparsity to gradients.
sparsity_optimizer.apply_sparsity()
# Model forward and backward as usual.
loss = model(...)
loss.backward()
# Update weights.
optimizer.step()
# Update sparsity pattern.
sparsity_optimizer.step()
SparsityOptimizer direct construction#
The above example is also equivalent to:
# Construct model and optimizer as usual
model = torch.nn.Module(...)
optimizer = cstorch.optim.SGD(...)
# Construct a sparsity optimizer using configuration helper.
sparsity_optimizer = cstorch.sparse.StaticSparsityOptimizer
[
(name, param)
for name, param in model.named_parameters()
if len(param.shape) > 1 and "emb" not in name
],
sparsity=0.7,
init_method="topk",
)
for step in dataloader:
# Ensure weights state are sparse. Also sets up gradient
# hook to apply sparsity to gradients.
sparsity_optimizer.apply_sparsity()
# Model forward and backward as usual.
loss = model(...)
loss.backward()
# Update weights.
optimizer.step()
# Update sparsity pattern.
sparsity_optimizer.step()
Checkpoint Format#
Unlike in previous versions of the Cerebras software, all weights always get
0.0 for the values that are pruned. If the sparsity patterns (as masks) are
needed for conversion to other sparse model implementations, they can be found
in the checkpoint under optimizer.sparsity.state.$param_name.mask. There is
no need to run a finalizer.py script to remove any sentinel values.
Implementation notes#
The Cerebras Wafer Scale Cluster implements sparsely natively in CSR (compressed sparse row) form, but for ease of use, all models are represented as dense + mask at the PyTorch level. The compiler takes care of automatically translating the representations behind the scenes.
Note, PyTorch does have mechanisms for both representing sparse tensors and utilities for pruning
networks.
However, sparse tensors require custom kernels and lower compatibility with
existing models and utilities. In particular, a torch.nn.Parameter can not
hold a torch.sparse.tensor without workarounds. The torch.prune
utilities are convenient, but the asynchronous and precompiled nature of
computation on the WSE requires a custom solution. Cerebras will attempt to
bridge compatibility with them in the future.